Note
Click here to download the full example code
Scalarized Multi-Objective Using ParEGO¶
This example builds on SVM with Cross-Validation.
Optimize both the final performance and the time used for training.
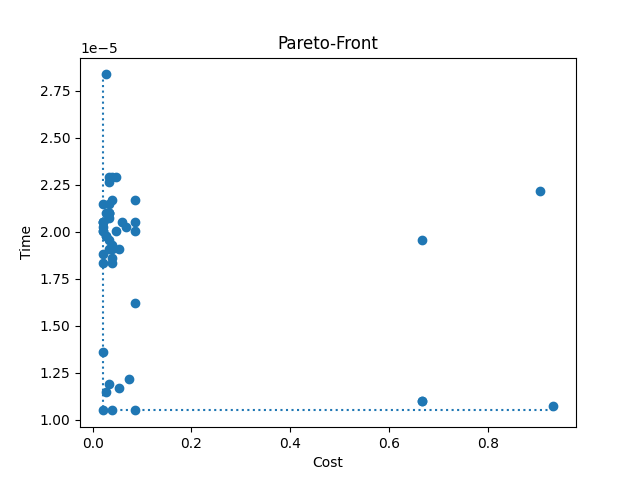
Out:
Default config's cost: 0.033333, training time: 0.000037 seconds
Optimizing! Depending on your machine, this might take a few minutes.
import logging
from smac.multi_objective.parego import ParEGO
logging.basicConfig(level=logging.INFO)
import time
import matplotlib.pyplot as plt
import numpy as np
from ConfigSpace.conditions import InCondition
from ConfigSpace.hyperparameters import (
CategoricalHyperparameter,
UniformFloatHyperparameter,
UniformIntegerHyperparameter,
)
from sklearn import datasets, svm
from sklearn.model_selection import cross_val_score
from smac.configspace import ConfigurationSpace
from smac.facade.smac_hpo_facade import SMAC4HPO
from smac.scenario.scenario import Scenario
from smac.utils.constants import MAXINT
__copyright__ = "Copyright 2021, AutoML.org Freiburg-Hannover"
__license__ = "3-clause BSD"
# We load the iris-dataset (a widely used benchmark)
iris = datasets.load_iris()
def is_pareto_efficient_simple(costs):
"""
Plot the Pareto Front in our 2d example.
source from: https://stackoverflow.com/a/40239615
Find the pareto-efficient points
:param costs: An (n_points, n_costs) array
:return: A (n_points, ) boolean array, indicating whether each point is Pareto efficient
"""
is_efficient = np.ones(costs.shape[0], dtype=bool)
for i, c in enumerate(costs):
if is_efficient[i]:
# Keep any point with a lower cost
is_efficient[is_efficient] = np.any(costs[is_efficient] < c, axis=1)
# And keep self
is_efficient[i] = True
return is_efficient
def plot_pareto_from_runhistory(observations):
"""
This is only an example function for 2d plotting, when both objectives
are to be minimized
"""
# find the pareto front
efficient_mask = is_pareto_efficient_simple(observations)
front = observations[efficient_mask]
# observations = observations[np.invert(efficient_mask)]
obs1, obs2 = observations[:, 0], observations[:, 1]
front = front[front[:, 0].argsort()]
# add the bounds
x_upper = np.max(obs1)
y_upper = np.max(obs2)
front = np.vstack([[front[0][0], y_upper], front, [x_upper, np.min(front[:, 1])]])
x_front, y_front = front[:, 0], front[:, 1]
plt.scatter(obs1, obs2)
plt.step(x_front, y_front, where="post", linestyle=":")
plt.title("Pareto-Front")
plt.xlabel("Cost")
plt.ylabel("Time")
plt.show()
def svm_from_cfg(cfg):
"""Creates a SVM based on a configuration and evaluates it on the
iris-dataset using cross-validation. Note here random seed is fixed.
It is a multi-objective tae, because we wish to trade-off the time to train
and the algorithm's final performance.
Parameters:
-----------
cfg: Configuration (ConfigSpace.ConfigurationSpace.Configuration)
Configuration containing the parameters.
Configurations are indexable!
Returns:
--------
Dict: A crossvalidated mean score (cost) for the svm on the loaded data-set and the
second objective; runtime
"""
# For deactivated parameters, the configuration stores None-values.
# This is not accepted by the SVM, so we remove them.
cfg = {k: cfg[k] for k in cfg if cfg[k]}
# And for gamma, we set it to a fixed value or to "auto" (if used)
if "gamma" in cfg:
cfg["gamma"] = cfg["gamma_value"] if cfg["gamma"] == "value" else "auto"
cfg.pop("gamma_value", None) # Remove "gamma_value"
t0 = time.time()
clf = svm.SVC(**cfg, random_state=42)
t1 = time.time()
scores = cross_val_score(clf, iris.data, iris.target, cv=5)
cost_value = 1 - np.mean(scores) # Minimize!
# Return a dictionary with all of the objectives.
# Alternatively you can return a list in the same order
# as `multi_objectives`.
return {"cost": cost_value, "time": t1 - t0}
if __name__ == "__main__":
# Build Configuration Space which defines all parameters and their ranges
cs = ConfigurationSpace()
# We define a few possible types of SVM-kernels and add them as "kernel" to our cs
kernel = CategoricalHyperparameter(
name="kernel",
choices=["linear", "rbf", "poly", "sigmoid"],
default_value="poly",
)
cs.add_hyperparameter(kernel)
# There are some hyperparameters shared by all kernels
C = UniformFloatHyperparameter("C", 0.001, 1000.0, default_value=1.0, log=True)
shrinking = CategoricalHyperparameter("shrinking", [True, False], default_value=True)
cs.add_hyperparameters([C, shrinking])
# Others are kernel-specific, so we can add conditions to limit the searchspace
degree = UniformIntegerHyperparameter("degree", 1, 5, default_value=3) # Only used by kernel poly
coef0 = UniformFloatHyperparameter("coef0", 0.0, 10.0, default_value=0.0) # poly, sigmoid
cs.add_hyperparameters([degree, coef0])
use_degree = InCondition(child=degree, parent=kernel, values=["poly"])
use_coef0 = InCondition(child=coef0, parent=kernel, values=["poly", "sigmoid"])
cs.add_conditions([use_degree, use_coef0])
# This also works for parameters that are a mix of categorical and values
# from a range of numbers
# For example, gamma can be either "auto" or a fixed float
gamma = CategoricalHyperparameter("gamma", ["auto", "value"], default_value="auto") # only rbf, poly, sigmoid
gamma_value = UniformFloatHyperparameter("gamma_value", 0.0001, 8, default_value=1, log=True)
cs.add_hyperparameters([gamma, gamma_value])
# We only activate gamma_value if gamma is set to "value"
cs.add_condition(InCondition(child=gamma_value, parent=gamma, values=["value"]))
# And again we can restrict the use of gamma in general to the choice of the kernel
cs.add_condition(InCondition(child=gamma, parent=kernel, values=["rbf", "poly", "sigmoid"]))
# Scenario object
scenario = Scenario(
{
"run_obj": "quality", # we optimize quality (alternatively runtime)
"runcount-limit": 50, # max. number of function evaluations
"cs": cs, # configuration space
"deterministic": True,
"multi_objectives": ["cost", "time"],
# You can define individual crash costs for each objective
"cost_for_crash": [1, float(MAXINT)],
}
)
# Example call of the function
# It returns: Status, Cost, Runtime, Additional Infos
def_value = svm_from_cfg(cs.get_default_configuration())
print("Default config's cost: {cost:2f}, training time: {time:2f} seconds".format(**def_value))
# Optimize, using a SMAC-object
print("Optimizing! Depending on your machine, this might take a few minutes.")
# Pass the multi objective algorithm and its hyperparameters
smac = SMAC4HPO(
scenario=scenario,
rng=np.random.RandomState(42),
tae_runner=svm_from_cfg,
multi_objective_algorithm=ParEGO,
multi_objective_kwargs={
"rho": 0.05,
},
)
incumbent = smac.optimize()
# pareto front based on smac.runhistory.data
cost = np.vstack([v[0] for v in smac.runhistory.data.values()])
plot_pareto_from_runhistory(cost)
Total running time of the script: ( 0 minutes 17.857 seconds)